SBW 2.10.0 Released
March 2nd, 2014: We are proud to announce the immediate availability of the Systems Biology Workbench 2.10.0 from SourceForge.
March 2nd, 2014: We are proud to announce the immediate availability of the Systems Biology Workbench 2.10.0 from SourceForge.
Researchers in quantitative systems biology make use of a large number of different software packages for modeling, analysis, visualization, and general data manipulation. The Systems Biology Workbench (SBW), is a software framework that allows heterogeneous application components-written in diverse programming languages and running on different platforms-to communicate and use each others' capabilities via a fast binary encoded-message system. Our goal was to create a simple, high performance, open-source software infrastructure which is easy to implement and understand. SBW enables applications (potentially running on separate, distributed computers) to communicate via a simple network protocol. The interfaces to the system are encapsulated in client-side libraries that we provide for different programming languages.
At the last count, there were over 250 different packages for simulating cellular networks (see www.sbml.org ). This proliferation of tools has resulted in a variety of capabilities and interfaces. Though welcome in many respects, this proliferation has resulted in two unwelcome side effects:
1. Each tool uses its own format, often undocumented, to store models. The result is that a model saved in one tool cannot be loaded into another. This obviously hinders the free exchange of models from one tool to another.
2. The second problem is that many of the tools duplicate each other's capabilities. Writing simulation tools takes time, and many of the projects are short-lived, which means that the authors are unable to develop the tools very far. As a result, many of the tools provide similar functionality. Unlike other software development communities, there is little tradition of code reuse in the system biology community. As a result, the community has seen much duplicated effort.
Model Interchange The first problem, that of model exchange, has been addressed by introducing a standard format for all tool writers to employ. This standard is called Systems Biology Markup Language (SBML) Along with CellML (www.cellml.org), the introduction of a standard format is beginning to make a significant impact on tools writers, and the majority of the most widely used tools now employ SBML as a means to exchange models.
Code Reuse The second issue is more difficult to address, that is how to encourage code reuse in the community. Our attempt to resolve this has been to develop a software framework called the System Biology Workbench. The workbench allows different tools to expose programmatic functionality to other tools. This means that a developer can now build on previous work without having to understand in detail the often intricate internal workings of other tools. All a developer need know is the interface that the tool exposes. Thus, a particular tool may expose a time-dependent simulation interface from a simulation tool, another tool developer-rather than invent another simulation tool-can exploit this capability and develop a new tool that can carry out additional functions. The workload for the second developer is greatly reduced, and they can instead concentrate on novel functionality.
This work is and was supported through the generous support of NIH, DARPA and the DOE
SBW consists of two components, a broker for routing messages and modules which send and receive messages. All connections between modules and a broker are via standard TCP/IP sockets. All messages are transmitted in a binary format for maximum performance.

If a message needs to be sent between two different computers, then messages are sent first to the broker on the remote machine, this in turn routes the message to the correct remote module. Modules may be written in a variety of languages, including, Java, C/C++, Delphi, Perl, Python and Matlab.
SBW messages come in four flavours:

Call/Send Message Format
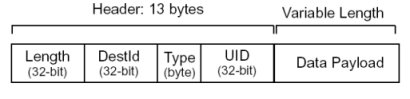
Reply Message Format
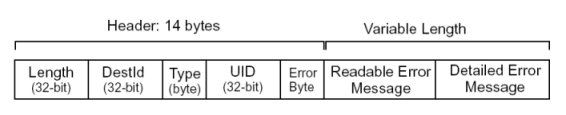
Error Message Format
Data Types
SBW supports seven basic data types:
byte, boolean, integer (32-bit), double (64 bit), string, array and list
Each data type is proceeded by a type type to indicate the type of data that follows. For example the data type byte, actually comprises of two bytes, one byte to indicate that the following type is a byte and the data byte itself. Boolean types are represented in exactly the same way as a byte type, the value of the byte when set to zero represents false and a value of one true.
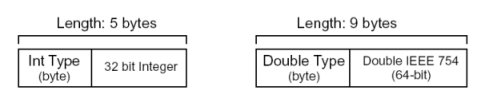
Integer and double data types have the following structures.
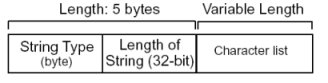
The string data type has the following structure. The string itself is made up of an unsigned 32-bit integer denoting the number of bytes in the string. The remainder of the data consists of the sequence of characters that make up the string which is also null terminated.
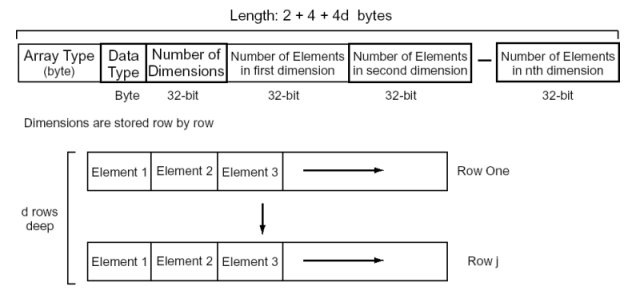
Arrays are multi-dimensional objects of arbitrary size containing homogeneous data. Arrays start with a header made up of one byte indicating the data type stored in the array, and an integer indicating the number of dimensions, followed by a sequence of integers, one for each dimension, denoting the number of elements in each dimension. The header is therefore (2 + 4 + 4d) bytes long, where d equals the number of dimensions of the array. Array access can be optimized at the module if it is known that the data type has a fixed size. This is especially the case for simple types such as integers and doubles. In these cases, the application can carry out block copies of the data in order to greatly improve performance. The array type has the following structure:
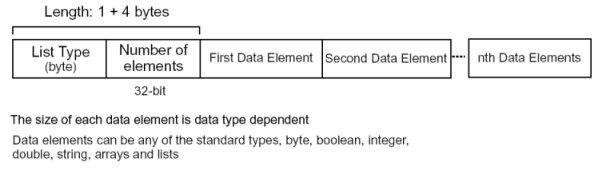
Lists are recursively defined structures for storing heterogeneous data. This means that lists can be used to store other lists which allows complex relationships to be represented. A list is a much simpler structure that an array. A list starts with a list type byte, followed by a 32-bit integer indicating the number of items in the list. Each items in the list can be any of the data types previously described, including a list. The list type has the following structure.
This simple binary protocoll has been implemented in language bindings available for every major programming language.
The Systems Biology Workbench Project was originally funded by a generous grant from the Japan Science and Technology Corporation through the ERATO Kitano Systems Biology Project. Currently support comes from the DARPA BioSPICE and Department of Energy GTL programs for which we an extremely grateful. The orignal authors of the SBW included Andrew Finney, Mike Hucka and Herbert Sauro with Hamid Bolouri, John Doyle and Hiroaki Kitano acting as principal investigators.
A detailed description of SBW can be found in our paper published in "Next generation simulation tools: the Systems Biology Workbench and BioSPICE integration."OMICS. 2003 Winter;7(4):355-72. Sauro, Hucka, Finney, Wellock, Bolouri, Doyle, Kitano.